How it works¶
Quick sum-up¶
As the name indicates SNAPPy is a pipeline, a series of tools used in succession or parallel to achieve an end. The general flow of the tool can be see in Fugure 1.
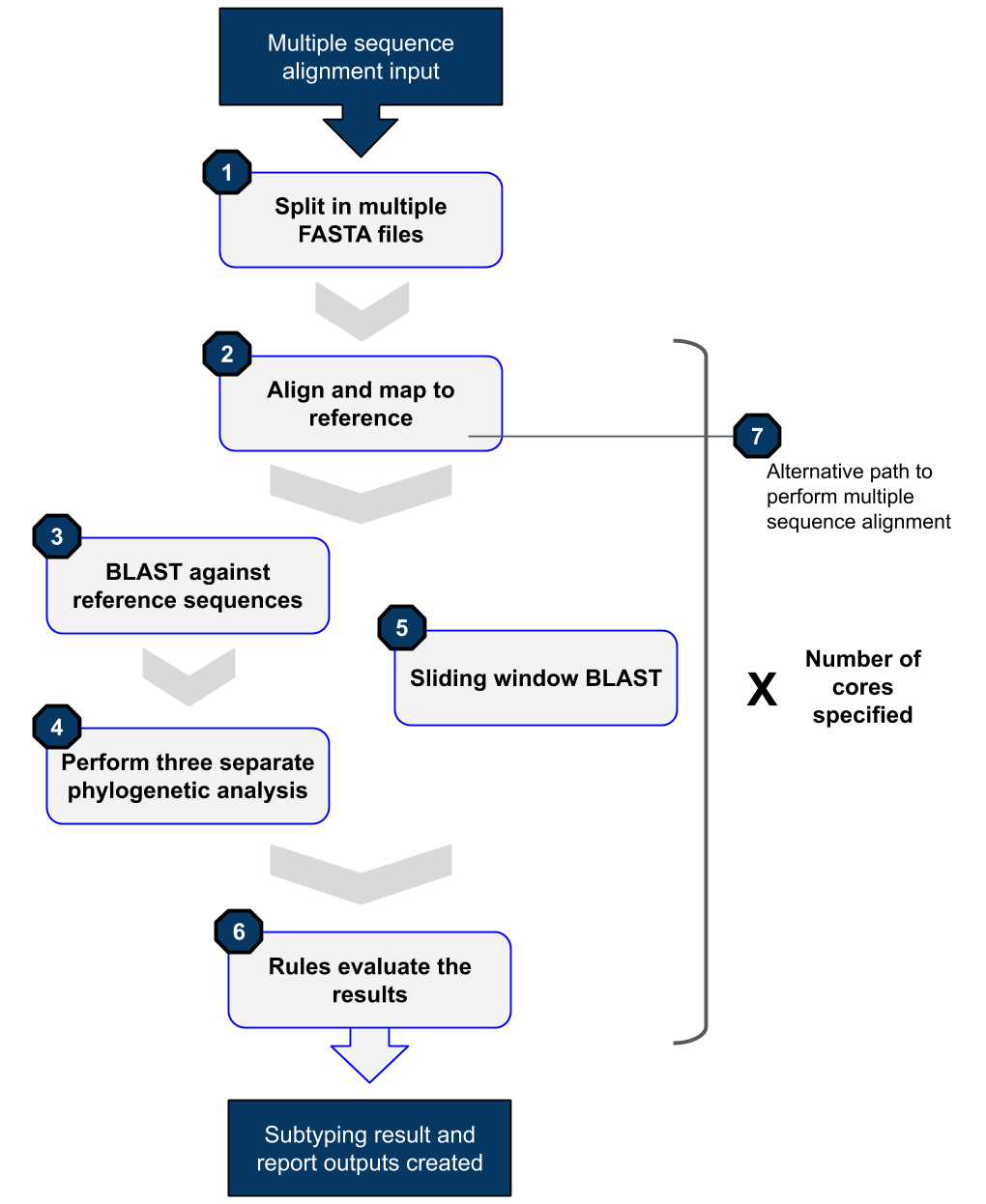
Figure 1: Schematic representation of the SNAPPy workflow.
- From a multiple sequence alignment a series of single FASTA sequence files are build.
- Each of these FASTA files is then aligned to the HIV-1 reference genome (HXB2 (K03455)).
- Each of the aligned files are them BLASted against a database of HIV-1 reference sequences (see Reference Sequences). The closest sequence according to the BLAST analysis is outputted in the ‘report_subtype_results.csv’ file in the column ‘closser_ref’.
- The BLAST results are them used to make 3 groups of references sequences of size 48: 1) all references; 2) only references of subtypes and sub-subtypes ; 3) only references of circulating recombination forms (CRF). - Each of these tree groups, together with the target sequence and an non-HIV-1 sequence (see Reference Sequences), are used to create 3 phylogenetic trees. These phylogenetic trees are then evaluated to see if the target sequence belong to a monophyletic clade with references of one, and only one, subtype or CRF. If this happens the subtype/CRF and the support values (Shimodaira-Hasegawa test) for the respective node are outputted in the ‘report_subtype_results.csv’ file in the columns ‘node_all_refs’, ‘s_node_all_refs’, ‘node_pure_refs’, ‘s_node_pure_refs’, ‘node_recomb_refs’, and ‘s_node_recomb_refs’.
- In parallel a Sliding window BLAST is performed against a database of HIV-1 reference sequences (see Reference Sequences), in order to look for evidence of recombination. The output from the sliding window BLAST can be seen in the column ‘recomb_result’ in the ‘report_subtype_results.csv’ file.
- These eight outputs are them combined and evaluated using a set of Decision Rules creating the final SNAPPy output, which can be seen in the column ‘result’ in both output files (‘subtype_results.csv’ and ‘report_subtype_results.csv’).
- SNAPPy can also be used to perform intermediate tasks of its pipeline, such as multiple sequence alignment (MSA). This characteristic is extremely useful for large scale HIV-1 alignment against a reference genome (HXB2 (K03455)). The MSA alignment created by SNAPPy is outputted to the file ‘all_aligned.fasta’.
In both subtyping outputs (‘subtype_results.csv’ and ‘report_subtype_results.csv’) there is also a column named ‘id’ which refers to the internal identifier used inside SNAPPy instead of the FASTA file header. If one wants to check the SNAPPys intermediate files first look for the connection between the ‘id’ that correspond to the desired FASTA in the ‘report_subtype_results.csv’ file.
If you desire to know more about how each of these steps are processed inside SNAPPy please read the following sections. If you want to use SNAPPy here is a list of the SNAPPy commands.
Reference Sequences¶
In all instances of SNAPPy the HXB2 (K03455) reference genome was used as a genomic position map reference, the HIV sequence database as a great visualization of this reference genome .
In the phylogenetic inferences the CONSENSUS_CPZ sequence from the HIV sequence database was used as outgroup for tree rooting (Alignment type: Consensus/Ancestral, Year: 2002, Organism: Other SIV, DNA/Protein: DNA, Region: genome, Subtype: ALL, Format: FASTA, Alignment ID : S02CG1).
We tried our best to create the best possible dataset of HIV-1 reference sequences. This task was extremely challenging. We based our dataset on previously curated data like the HIV sequence database 2010 subtype reference genomes sequences compendium and the HIV Drug Resistance Database reference sequences . Our goal was to create a comphresensive yet not too extensive dataset of HIV-1 references. The subtype reference dataset used in SNAPPy at the moment consists of 493 genomic sequences, and can be downloaded here , this dataset was applied in the task BLAST. A subset (n= 56) of this reference dataset that does not contai CRFs references ( with the exception of 01 and 02) was applied in the sliding window BLAST task, and can be downloaded from here. Following there are the lists of these two groups of HIV-1 subtype references (Nomenclature: ‘group/subtype/sub-subtype/CRF’-’Accession number’):
- Complete dataset of reference sequences (n = 491) used in BLAST:
A1-DQ676872, A1-AB253421, A1-AB253429, A2-AF286238, A2-GU201516, A2-AF286237, B-K03455, B-AY423387, B-AY173951, B-AY331295, C-U52953, C-U46016, C-AF067155, C-AY772699, D-K03454, D-AY371157, D-AY253311, D-U88824, F1-AF077336, F1-AF005494, F1-AF075703, F1-AJ249238, F2-AY371158, F2-AJ249236, F2-AJ249237, F2-AF377956, G-AF084936, G-AF061641, G-U88826, G-AY612637, H-AF190127, H-AF190128, H-AF005496, H-FJ711703, J-EF614151, J-GU237072, J-AF082394, K-AJ249235, K-AJ249239, 01_AE-GQ477441, 01_AE-GU564221, 01_AE-U54771, 02_AG-AY271690, 02_AG-AB485636, 02_AG-L39106, 03_AB-AF193276, 04_cpx-AF049337, 04_cpx-AF119820, 04_cpx-AF119819, 05_DF-AF076998, 05_DF-AF193253, 05_DF-AY227107, 06_cpx-AF064699, 06_cpx-AY535659, 06_cpx-AB286851, 07_BC-EF368372, 07_BC-EF368370, 07_BC-AF286230, 08_BC-HM067748, 08_BC-AY008715, 09_cpx-AJ866553, 09_cpx-AY093605, 09_cpx-AY093603, 09_cpx-AY093607, 10_CD-AF289548, 10_CD-AF289549, 10_CD-AF289550, 11_cpx-AF492624, 11_cpx-AF492623, 11_cpx-AJ291718, 12_BF-AF408629, 12_BF-AF408630, 12_BF-AF385936, 13_cpx-DQ845388, 13_cpx-DQ845387, 13_cpx-AF460972, 14_BG-AF450096, 14_BG-AF450097, 14_BG-GU230137, 15_01B-DQ354120, 15_01B-AF516184, 15_01B-AF530576, 16_A2D-AY945736, 16_A2D-AF286239, 17_BF-EU581825, 17_BF-EU581827, 17_BF-EU581828, 18_cpx-AF377959, 18_cpx-AY586541, 18_cpx-AY894993, 19_cpx-AY588971, 19_cpx-AY588970, 19_cpx-AY894994, 20_BG-AY586545, 21_A2D-AY945737, 21_A2D-AF457051, 21_A2D-AF457072, 22_01A1-AY371159, 22_01A1-GQ229529, 23_BG-AY900571, 23_BG-AY900572, 24_BG-AY900574, 24_BG-AY900575, 24_BG-FJ670526, 25_cpx-EU693240, 25_cpx-EU697906, 25_cpx-EU697908, 26_A5U-FM877780, 26_A5U-FM877782, 26_A5U-FM877777, 27_cpx-AJ404325, 27_cpx-AM851091, 28_BF-DQ085872, 28_BF-DQ085873, 28_BF-DQ085874, 29_BF-DQ085876, 29_BF-AY771590, 29_BF-DQ085871, 31_BC-EF091932, 31_BC-AY727526, 31_BC-AY727527, 32_06A6-AY535660, 33_01B-AB547464, 33_01B-DQ366659, 33_01B-DQ366662, 34_01B-EF165541, 35_AD-EF158043, 35_AD-EF158040, 35_AD-EF158041, 36_cpx-EF087995, 36_cpx-EF087994, 37_cpx-EF116594, 37_cpx-AF377957, 38_BF-FJ213781, 38_BF-FJ213782, 38_BF-FJ213780, 39_BF-EU735534, 39_BF-EU735536, 39_BF-EU735535, 40_BF-EU735538, 40_BF-EU735540, 40_BF-EU735539, 42_BF-EU170155, 43_02G-EU697904, 43_02G-EU697907, 43_02G-EU697909, 44_BF-FJ358521, 45_cpx-FN392874, 45_cpx-FN392876, 45_cpx-FN392877, 46_BF-DQ358801, 46_BF-DQ358802, 46_BF-HM026456, 47_BF-GQ372987, 47_BF-FJ670529, 49_cpx-HQ385477, 49_cpx-HQ385479, 49_cpx-HQ385478, O-L20587, O-L20571, O-AY169812, O-AJ302647, N-AY532635, N-AJ006022, N-AJ271370, P-HQ179987, P-GU111555, 42_BF-EU170142, 42_BF-EU170151, 42_BF-EU170147, 03_AB-AF193277, 03_AB-AF414006, 44_BF-AY536235, 20_BG-AY586544, 20_BG-DQ020274, 32_06A6-DQ167215, 34_01B-EF165539, 34_01B-EF165540, 48_01B-GQ175881, 48_01B-GQ175882, 48_01B-GQ175883, 32_06A6-KM606632, 20_BG-KT276270, 41_CD-KX907411, 41_CD-KX907417, 41_CD-KX907430, 03_AB-MF109476, 50_A1D-JN417241, 51_01B-JN029801, 52_01B-AY945734, 53_01B-JX390610, 54_01B-JX390977, 55_01B-JX574663, 56_cpx-KC852174, 57_BC-KC899008, 58_01B-KC522031, 59_01B-KC462190, 60_BC-KC899079, 61_BC-KC990124, 62_BC-KC870037, 63_02A-JX500706, 64_BC-KC870043, 65_cpx-KC870030, 67_01B-KC183779, 68_01B-KC183782, 69_01B-LC027100, 70_BF-KJ849809, 70_BF-KJ849761, 71_BF-DQ358811, 71_BF-KJ849775, 72_BF-KJ671533, 72_BF-KJ671537, 73_BG-KM248765, 74_01B-KR019770, 78_cpx-KU161143, 53_01B-JX390611, 53_01B-JX390612, 63_02A-KJ197201, 77_cpx-KX673818, 52_01B-DQ366664, 54_01B-JX390976, 57_BC-KC870044, 59_01B-KJ484433, 60_BC-KC899081, 62_BC-KC870034, 67_01B-KC183780, 69_01B-AB845349, 74_01B-KR019771, 78_cpx-KU161145, 50_A1D-JN417240, 51_01B-KJ485697, 55_01B-KF927150, 56_cpx-KC852173, 58_01B-KC522033, 61_BC-KC990126, 64_BC-KC898994, 65_cpx-MH051841, 68_01B-KF758551, 73_BG-AY882421, 77_cpx-KX673820, 63_02A-JX500705, 63_02A-JX500700, 63_02A-JX500699, 63_02A-JN230353, 63_02A-KJ197200, 63_02A-KJ197202, 63_02A-JX500704, 70_BF-KJ849758, 70_BF-KJ849762, 71_BF-KJ849760, 71_BF-KJ849769, 71_BF-KJ849771, 71_BF-KJ849777, 71_BF-KJ849778, 71_BF-KT427816, 72_BF-KJ671534, 72_BF-KJ671535, 72_BF-KJ671536, 79_0107-KY216146, 79_0107-KY216147, 79_0107-KY216148, 80_0107-MH843712, 80_0107-MH843713, 81_cpx-MH986016, 81_cpx-MH986017, 82_cpx-KU820825, 82_cpx-KU820831, 82_cpx-KU820836, 82_cpx-KU820837, 82_cpx-KU820844, 82_cpx-KU820845, 83_cpx-KU820823, 83_cpx-KU820824, 83_cpx-KU820826, 83_cpx-KU820828, 83_cpx-KU820829, 83_cpx-KU820833, 83_cpx-KU820834, 83_cpx-KU820839, 83_cpx-KU820842, 83_cpx-KU820843, 83_cpx-KU820847, 85_BC-KU992928, 85_BC-KU992929, 85_BC-KU992930, 85_BC-KU992931, 85_BC-KU992932, 85_BC-KU992934, 85_BC-KU992935, 85_BC-KU992936, 85_BC-KU992937, 86_BC-KX582251, 86_BC-KX582250, 86_BC-KX582249, 87_cpx-KC899012, 87_cpx-KC898992, 87_cpx-KF250408, 88_BC-KC898979, 88_BC-KC898975, 88_BC-KF250402, 90_BF1-KY628225, 90_BF1-KY628223, 90_BF1-KY628221, 90_BF1-KY628219, 90_BF1-KY628218, 90_BF1-KY628216, 92_C2U-MF372648, 92_C2U-MF372647, 92_C2U-MF372645, 93_cpx-MF372651, 93_cpx-MF372649, 93_cpx-MF372646, 94_cpx-MH141491, 94_cpx-MH141492, 94_cpx-MH141493, 94_cpx-MH141494, 96_cpx-KF850149, 96_cpx-MG518477, 96_cpx-MG518476, 99_BF-MH986013, 99_BF-MH986014, A1-AF069670, A1-M62320, A1-AF484509, A1-EF545108, C-AF443091, H-KU310618, 11_cpx-AY371152, 11_cpx-AJ291720, 16_A2D-AF457060, 22_01A1-GQ229530, 25_cpx-DQ826726, 26_A5U-FM877778, 44_BF-AY536238, 45_cpx-FN392875, 01_AE-KP411840, 01_AE-KP411841, 01_AE-KP718930, 02_AG-AB052867, 02_AG-AJ239083, 02_AG-AJ508595, 02_AG-AY444811, 02_AG-FJ388896, 02_AG-FJ694791, 02_AG-JF683786, 02_AG-JF683795, 02_AG-JN248585, 02_AG-KU749413, 02_AG-KM606636, 02_AG-KP411843, 02_AG-KP411844, 02_AG-KR067668, 02_AG-KT124792, 06_cpx-KU168301, 06_cpx-KX389609, 09_cpx-AJ866556, 11_cpx-AJ291719, 11_cpx-AY371151, 11_cpx-JF683802, 11_cpx-KX389633, 18_cpx-AY371166, 19_cpx-AY894995, 25_cpx-KY392772, A1-AF069669, A1-AF069671, A1-AF457068, A1-AY521629, A1-AB287378, A1-DQ396400, A1-AM000053, A1-EU861977, A1-FJ388893, A1-FJ670519, A1-KU749409, A1-KT152839, A1-KX232613, B-AF362994, B-AF005495, B-AY795904, B-AY682547, B-DQ207940, B-FJ460499, B-DQ358806, B-DQ383750, B-DQ383752, B-AB221125, B-DQ396398, B-EF637049, B-EF637050, B-EF637054, B-EF637056, B-FJ195091, B-FJ388890, B-HM030559, B-GQ372988, B-EU839596, B-EU839600, B-EU839606, B-JF320008, B-JF320144, B-JF683738, B-JF683796, B-JF683801, B-HQ215554, B-JN235958, B-JN251896, B-JN692432, B-JN692433, B-JN692439, B-JN692448, B-JN692455, B-JN692460, B-JN692467, B-JF804810, B-JF804812, B-JF804813, B-KY658689, B-KF384810, B-KJ849785, B-KJ849804, B-KJ849808, B-KJ849814, B-KJ849780, B-KP411822, B-KY465969, B-KT427650, B-KT427671, B-KT427681, B-KT427707, B-KT427720, B-KT427735, B-KT427746, B-KT427757, B-KT427811, B-KT427828, B-KT124761, C-AF286223, C-AF286227, C-AF286233, C-AF286234, C-AF457061, C-AY255823, C-AY255824, C-AY255825, C-DQ207941, C-AY734554, C-AB286849, C-EU786673, C-FJ388901, C-GQ999983, C-KU749412, C-KU749430, C-KP411830, C-KP411834, C-KY496624, C-KT022371, C-KT124786, C-KU319528, C-KU319529, C-KU319551, C-KX907353, C-KY392767, D-AF442569, D-AF484514, D-AY773338, D-AY795907, D-AY444799, D-DQ054367, D-DQ912823, D-FJ388945, D-KU749394, D-KU168272, D-KX907406, D-KY392769, F1-DQ189088, F1-FJ900266, F1-AB485658, F1-GQ290462, F1-JX140671, F1-KY392770, F2-JX140672, G-DQ168573, G-AB231893, G-AY586547, G-JN106043, G-KJ948662, G-KU168277, H-KU168273, H-KY392777, J-KU168280, J-KU310620
- Dataset of reference sequences (n = 56) used in the sliding window BLAST
01_AE-GQ477441, 01_AE-GU564221, 01_AE-U54771, 01_AE-KP411840, A1-DQ676872, A1-AB253421, A1-AB253429, A2-AF286238, A2-GU201516, A2-AF286237, B-K03455, B-AY423387, B-AY173951, B-AY331295, C-U52953, C-U46016, C-AF067155, C-AY772699, D-K03454, D-AY371157, D-AY253311, D-U88824, F1-AF077336, F1-AF005494, F1-AF075703, F1-AJ249238, F2-AY371158, F2-AJ249236, F2-AJ249237, F2-AF377956, G-AF084936, G-AF061641, G-U88826, G-AY612637, H-AF190127, H-AF190128, H-AF005496, H-FJ711703, J-EF614151, J-GU237072, J-AF082394, K-AJ249235, K-AJ249239, O-L20587, O-L20571, O-AY169812, O-AJ302647, N-AY532635, N-AJ006022, N-AJ271370, P-HQ179987, P-GU111555, 02_AG-AB485636, 02_AG-AY271690, 02_AG-L39106, 02_AG-JF683786
Alignment to reference¶
After splitting the MSA in several FASTA files each of them is aligned to the HIV-1 reference genome (HXB2 (K03455)). The module SeqIO from Biopython is used to parse and manipulate the FASTA files in SNAPPy. The alignment is done using MAFFT v7.245. The alignment method used does not allow any gaps in the reference sequence (comand: ‘mafft –quiet –add “target” –keeplength “reference”). After the alignment is performed the target sequence is trimed to only contain the genomic region specified by the user in the ‘config.yaml’ file. Being the currently available options ‘GAG’, ‘PR’, ‘RT’, ‘PR-RT’, ‘INT’, ‘POL’, ‘ENV’, and ‘GAG-POL-ENV’. The resulting file is them written to the folder ‘aligned’ with the following notation: aligned_{internal_id}.fasta.
BLAST¶
The curated and aligned sequence files obtained are them BLASTed against a local database of HIV-1 reference sequences (see Reference Sequences). For this task BLAST v2.7.1 is used (comand; ‘blastn -db “references” -query “target” -out “blast_output” -word_size 10 -outfmt 10 -evalue 1.e-10’). The BLAST parameters ‘word_size’ = 10 and “evalue” cutoff = 1.e-10 showed good and consistent performance for the case of highly similar sequences. The results were sorted by bitscore ( higher is better). The BLAST result with the best score is outputted in the ‘report_subtype_results.csv’ file in the column ‘closser_ref’. The BLAST results are also used to make 3 groups of references sequences: containing the first 48 results; containing the first 48 results of only subtype references; containing the first 48 results of only CRF references. These three groups of reference sequences are then used in the phylogenetic analysis (see Phylogenetic inference. The intermediate files of the BLAST analysis are outputted to the folder ‘blast’ with the following notation: blast_{internal_id}.txt. Please notice that for the split in subtype and CRF references in this step of SNAPPy CRFs 01 and 02 are treated as subtypes an not CRFs. These decision was made based on the high prevalence of this CRFs and their ambiguous origin (Gao F, et al. J Virol. 1996; Abecasis AB, el al. J. Virol. 2007)
Phylogenetic inference¶
The target sequence (see Alignment to reference) and a non-HIV-1 sequence (for rooting, see Reference Sequences) are added to each of the three previously selected groups of 48 references (see BLAST). Groups of 50 sequences showed to be a contained and yet comprehensive set of sequences to perform the phylogenetic inference. To perform the phylogenetic analysis IQ-TREE v1.6.9 was used (comand: iqtree -s “msa_50_seqs” -quiet -m GTR+F+G4 -fast -alrt 1000 -seed 0). The ETE toolkit is used to parse and manipulate the phylogenetic trees created in SNAPPy. After rooting on the outgroup it is inferred if the target sequence is in a monophyletic clade with references of a same subtype/CRF. If that happens the subtype/CRF of those references is outputted together with the support values for that node (Shimodaira-Hasegawa test, as implemented in IQ-TREE). Resulting in six output columns in the ‘report_subtype_results.csv’ file: ‘node_all_refs’, ‘s_node_all_refs’, ‘node_pure_refs’, ‘s_node_pure_refs’, ‘node_recomb_refs’, and ‘s_node_recomb_refs’. The intermediate files of the phylogenetic analysis are outputted to the folder ‘trees’ with the following notation: {type}_{internal_id}.nwk (being {type} ‘all’, ‘pure’, and ‘recomb’ referring to the set of references used for that phylogenetic reconstruction).
Sliding window BLAST¶
This step of the pipeline starts from the sequence files created in the Alignment to reference section. The positions with gaps (‘-‘) in the target sequence are excluded. The length of the sliding window is 400 nucleotides. Fragments with length inferior to 400 will not be processed by this approach, and ‘impossible to test recomb (lenght < 400bp)’ will be written to the output file. The step size is 50 nucleotides, which creates 8 bins for each window. The result for each BLAST window is the subtype of the top result (bitscore). If more than one sequence of different subtypes have the same top score the output for all bins of that window is null. If the BLAST fails or no output is produced the output for all bins of that window is null. At the end of all sliding windows being processes several bins may have multiple outputs, a majority rule is applied to decide the final subtype for that bin. In case of tie the result for that bin is null. In this sliding window BLAST we are only BLASting against a database of HIV-1 subtype references, plus the CRFs 01 and 02 due to the reasons previously discussed in BLAST. The BLAST command used for each window is : ‘blastn -db “database” -query “target -out’, “output_file_name” -word_size 10 -outfmt 10 -evalue 1.e-100’. The database file for the BLAST can be consulted in this link . Similarly to what was mentioned in BLAST section these parameters proved to be very useful in the case of highly similar sequences like HIV-1. Please consult Figure 2 for a graphic explanation of this process. The resulting file is them written to the folder ‘blast’ with the following notation: recblast_{internal_id}.txt.
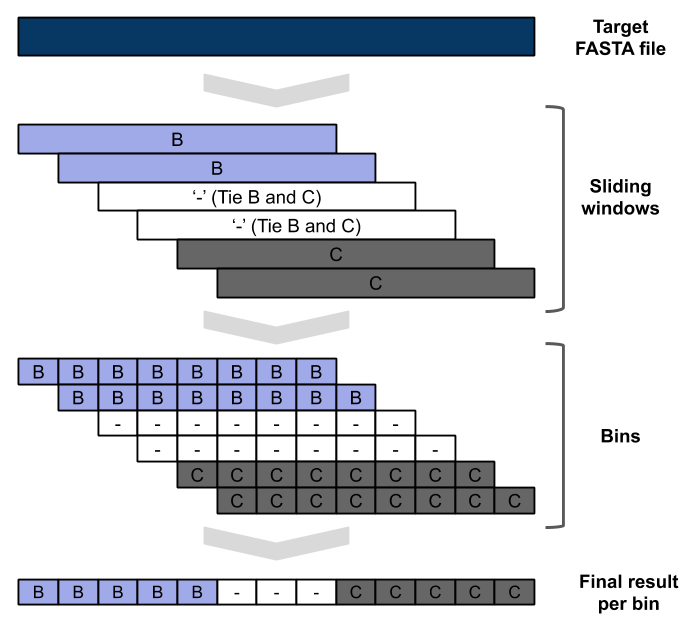
Figure 2: Schematic representation of the sliding window approach. Demonstrating evidence of B/C recombination.
Decision Rules¶
Given the previously described outputs produced by SNAPPY a set of decision rules were built. The rules are executed in the order they are shown. This means that for instance rule p2 is only applied after the requirements were not meet for rules p1 and c1. To simplify the rules description we describe the results from the Sliding window BLAST as follows: no recombination - if only one subtype/CRF is outputted; simple recombination - if two subtypes/CRFs are outputted, complex recombination - if more than two subtypes/CRFs are outputted. Tree ‘all’, ‘pure’, and ‘recomb’ refer, respectively, to the results from the phylogenies with all closest reference sequences, only subtype closest reference sequences, and only CRF closest reference sequences. Good support refers to Shimodaira-Hasegawa test ( as implemented in IQ-TREE) result >= 0.9, while a great support refers to results >= 0.95. This nomenclature has as only objective simplifying the description of the following rules and should not be applied outside this context:
Rules:
- p1: All analysis report the same output. Output: all tree result;
- c1: Simple recombination and all tree; recomb tree; and closer reference are equal. Both trees support is good. Output: all tree result;
- p2: All tree; pure tree; and recomb result are equal. Both trees support is good. Output: all tree result;
- p3: All tree; pure tree; and closer reference are equal. Both trees support is good. Output: all tree result;
- c2: All tree; recomb tree; and closer reference are equal. Both trees support is good. Output: all tree result;
- p4: Pure tree; closer reference; and recomb result are equal. Tree support is great. Output: pure tree result;
- c3: Recomb tree; closer reference; and recomb is simple. Tree support is great. Output: recomb tree result;
- b1: All tree; closer reference; and recomb result are equal. Tree support is great. Output: all tree result;
- There is evidence of complex recombination:
- c4: All tree and recomb tree give same result which is a CRF. Both trees support is good. Output: all tree result;
- p5: All tree; pure tree; and closer reference are equal. At least one of the trees has great support. Output: all tree result;
- c5: All tree; recomb tree; and closer reference give same result which is a CRF. Output: all tree result;
- p6: All tree; pure tree; and closer reference are equal. Output: all tree result;
- u1: The remaining cases with evidence of complex recombination. Output: ‘URF_CPX’;
- There is evidence of simple recombination:
- c6: All tree and recomb tree give same result which is a CRF. Both trees support is good. Output: all tree result;
- p7: All tree; pure tree; and closer reference are equal. At least one of the trees has great support. Output: all tree result;
- c7: All tree; recomb tree; and closer reference give same result which is a CRF. Output: all tree result;
- p8: All tree; pure tree; and closer reference are equal. Output: all tree result;
- u2: Remaining cases. Output: URF between the two results of the sliding window blast result;
- p9: All tree equal pure tree. Both trees support is good. Output: all tree result;
- If there is missing data regarding the recombination test:
- f1: If there is no result in the sliding window blast. Output: closer reference result;
- f2: If both the closer reference and sliding window blast results are missing but all tree agrees with pure tree result: Output: all tree result;
- f3: If both the closer reference and sliding window blast results are missing but all tree agrees with recomb tree result. Output: all tree result;
- f4: Remaining cases. Output: ‘impossible_to_determine’;
- f5: There is no evidence or recombination and the sliding window blast result is not null: Output sliding window blast result.
Other¶
Some intermediate files produced by SNAPPy are deleted before the end of the process. This was done to avoid waisting unnecessary disk space and at the same time simplify the user experience. However, all the intermediate files created by SNAPPy that may contain useful information regarding the analysis and the decisions made by the pipeline are kept.
Please keep in mind that SNAPPy does several analysis and some of them produce a large amount of outputs. If you are using SNAPPy for large scale analysis please understand that a large portion of disk space may be needed. This is a tradeof between transparency and computational resources that we thought may be the best for the user. At the end of each SNAPPy run it will delete an snakemake hidden folder named ‘.snakemake’ that occupies substantial amount of space. However, this folder contains all the logs about the tasks performed and may be useful for debugging. To change that behavior two lines need to be commented out in the ‘Snakefile’ (‘onsuccess:’ ‘shutil.rmtree(“.snakemake”)’), if you decide to do so do it at your own risk.